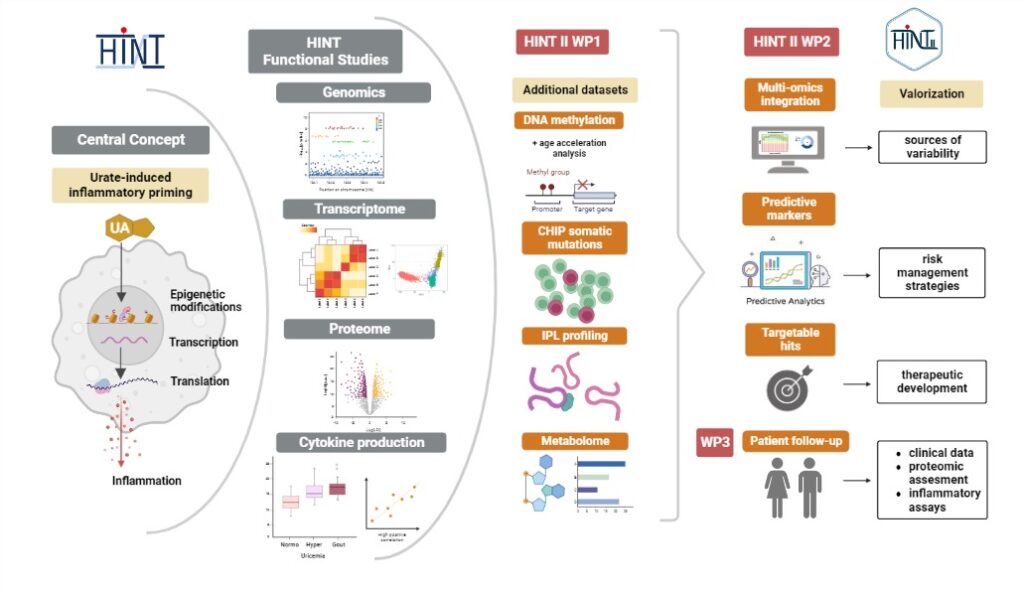
The deposition of uric acid monosodium crystals in joints induces inflammation, which is classical for gout. However, a growing body of evidence suggests that increased concentrations of uric acid also contribute to other inflammatory/metabolic diseases (e.g. metabolic syndrome, atherosclerosis). The mechanisms through which uric acid enhances the inflammatory setpoint of the immune system are unknown. The current project aims to investigate patients with gout, asymptomatic hyperuricemia, and controls. During the 36 months of this project, an innovative approach will be developed by analyzing new processes such as clonal hematopoiesis with undetermined potential, DNA methylation and age acceleration, the role of immune gene priming long non-coding RNA (immune gene priming long non-coding RNA) in the stability of epigenetic effects and the interaction of metabolic factors in the prediction of the inflammatory response. This will be realized through several scientific aims, including integrated multi-omics studies and comprehensive analysis which will allow the development of new prediction tools to identify people at risk of distant inflammatory events (gout attacks, cardiovascular events). DNA methylation data will be used to assess accelerated aging in correlation with urate levels and urate induced inflammatory priming. This kind of strategy will help to identify feature combinations that can stratify patients into clusters (e.q. fast and slow progression from
hyperuricemia to gout, CVD complications, accelerated aging). Also, a comprehensive systems biology approach to integrate large-scale genomic, transcriptomic and cytokine production data will be used to describe the baseline heterogeneity of immunological parameters, to identify inter-correlated immune components, infer functional connections within the immune system and build predictive models of cytokine-production capacity and trained immunity features in response to urate. Therefore, the project will progress from identified associations and mechanisms to functional validations and translation of results to clinical practice.